Introduction
In this tutorial, we will use the Recurrent Feature Elimination (RFE) method to select attributes in the Pima Indians Diabetes dataset. For this purpose the library caret will be used.
First we will split the dataset in train and validation datasets.
# ensure the results are repeatable
set.seed(1234)
# load the library
library(mlbench)
library(caret)
# load the data
data(PimaIndiansDiabetes)
data = PimaIndiansDiabetes
# split into training and validation datasets
set.seed(1234)
ind <- sample(2, nrow(data), replace=TRUE, prob=c(0.7, 0.3))
trainData <- data[ind==1,]
validationData <- data[ind==2,]
trainData <- trainData[complete.cases(trainData),]
validationData <- validationData[complete.cases(validationData),]Next we will use the rfe method of the caret package, setting up using the rfeControl method.
# define the control using a random forest selection function
control <- rfeControl(functions=nbFuncs, method="cv", number=10)
# run the RFE algorithm
results <- rfe(trainData[,1:8], trainData[,9], sizes=c(1:8), rfeControl=control)Let’s see the results.
# summarize the results
print(results)##
## Recursive feature selection
##
## Outer resampling method: Cross-Validated (10 fold)
##
## Resampling performance over subset size:
##
## Variables Accuracy Kappa AccuracySD KappaSD Selected
## 1 0.7453 0.3681 0.04390 0.12449
## 2 0.7546 0.4182 0.04598 0.12317
## 3 0.7715 0.4707 0.05759 0.14388 *
## 4 0.7584 0.4479 0.04145 0.10478
## 5 0.7546 0.4457 0.04429 0.10095
## 6 0.7490 0.4344 0.04393 0.09394
## 7 0.7509 0.4290 0.04686 0.11122
## 8 0.7491 0.4201 0.03507 0.07767
##
## The top 3 variables (out of 3):
## glucose, age, mass# list the chosen features
predictors(results)## [1] "glucose" "age" "mass"# plot the results
plot(results, type=c("g", "o"))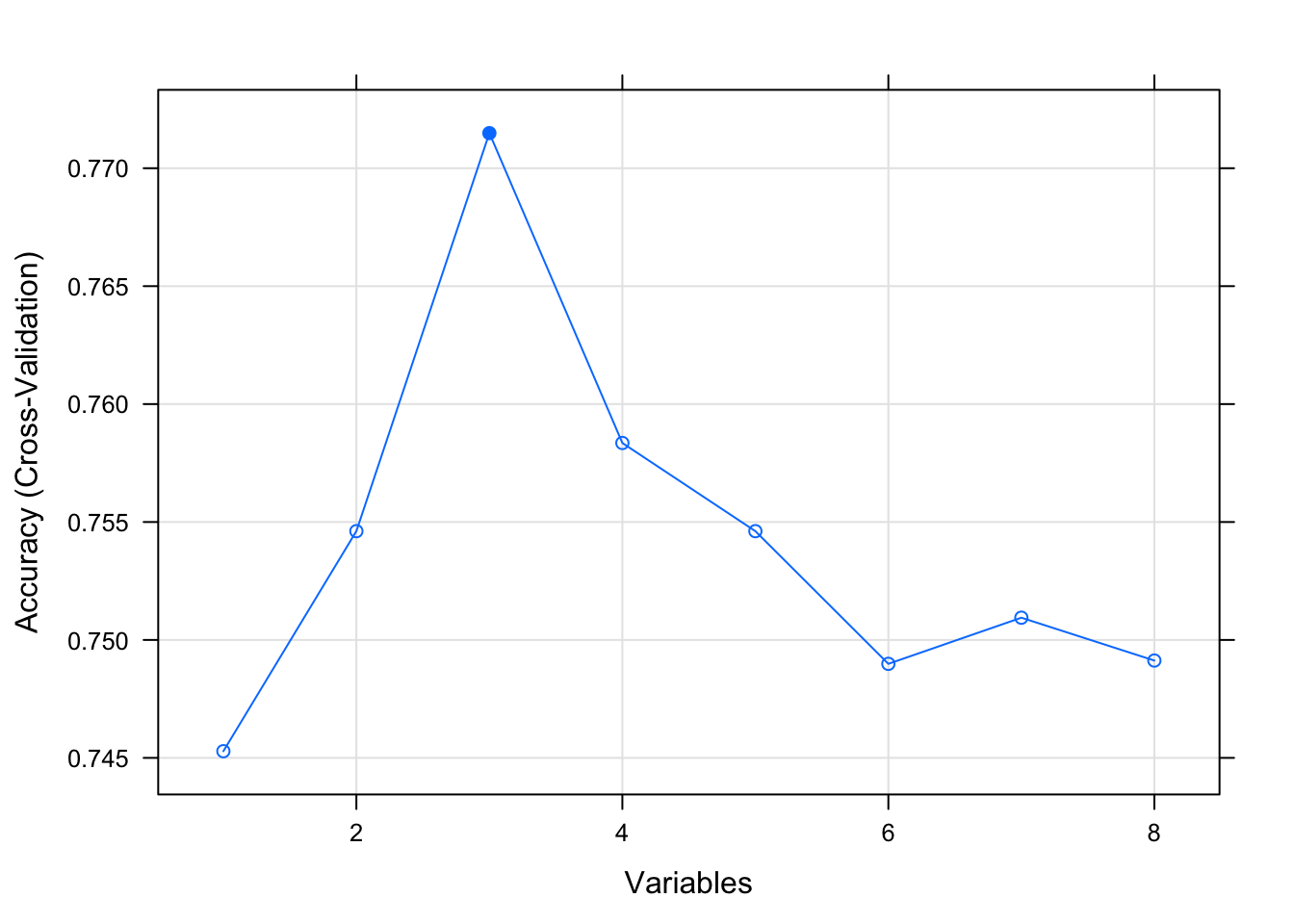
Naive Bayes
We will use the 3 top variables that come out of RFE in the Naive Bayes algorithm:
library(e1071)
(f <- as.formula(paste("diabetes", paste(results$optVariables, collapse=" + "), sep=" ~ ")))## diabetes ~ glucose + age + massmodel <- naiveBayes(diabetes ~ ., data=trainData, laplace = 1)
simpler_model <- naiveBayes(f, data=trainData, laplace = 1)
pred <- predict(model, validationData)
simpler_pred <- predict(simpler_model, validationData)
library(MLmetrics)
train_pred <- predict(model, trainData)
train_simpler_pred <- predict(simpler_model, trainData)
paste("Accuracy in training all attributes",
Accuracy(train_pred, trainData$diabetes), sep=" - ")## [1] "Accuracy in training all attributes - 0.760299625468165"paste("Accuracy in training RFE attributes",
Accuracy(train_simpler_pred, trainData$diabetes), sep=" - ")## [1] "Accuracy in training RFE attributes - 0.764044943820225"paste("Accuracy in validation all attributes",
Accuracy(pred, validationData$diabetes), sep=" - ")## [1] "Accuracy in validation all attributes - 0.764957264957265"paste("Accuracy in validation RFE attributes",
Accuracy(simpler_pred, validationData$diabetes), sep=" - ")## [1] "Accuracy in validation RFE attributes - 0.799145299145299"Using a simpler formula with 5 out of 8 attibutes we were able to get better generalization results in the validation dataset.